





Welcome to the Chromatin Epigenetics Lab

mviji@sastra.edu
Epigenetic Tools developed in the lab
NAM: Nucleosome Array Modifier
CED: Cancer Epigenetics Database
Nuclear Phenotype Classifier
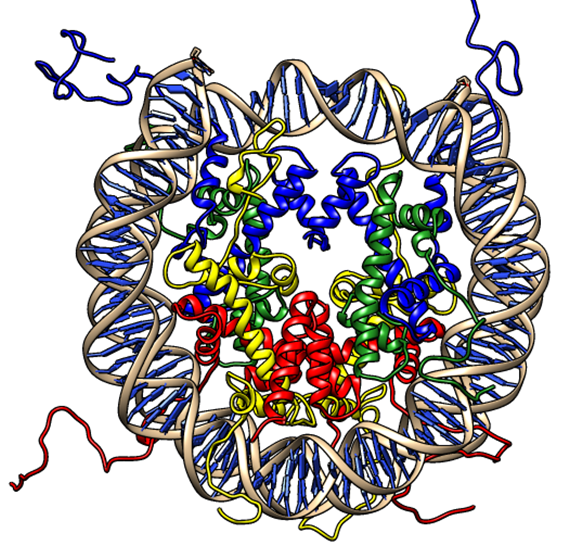
DNA in eukaryotic cell nuclei is packed and presented in the form of chromatin. The exquisite machinery involved in packing the metre long DNA within the nuclear confines (microns) provides a major challenge to regulatory molecules to access the DNA. Chromatin remodelling enzymes, covalent modifications of the histone tails, histone variants and several other factors influence transcription through chromatin. Reversible post translational modifications such as acetylation, methylation, phosphorylation, ubiquitination on the histone tails dictate gene expression patterns. Alterations in these profiles critically influence cellular differentiation, phenotypic variability, disease onset and progression. Emerging evidences strongly link abnormalities in histone modifications to autoimmune, inflammatory and neoplastic disorders and to neurological deficits.
Our laboratory is interested in understanding epigenetic signalling in disease and during apoptosis.We use biochemical assays, live cell imaging, histopathology and high throughput microscopy to dissect the role of histone modifications (on the core and linker histones) during the onset of diseases and their progression.
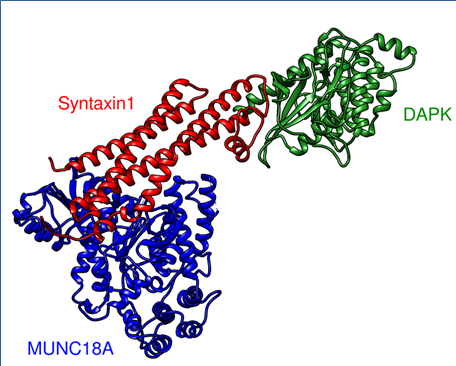
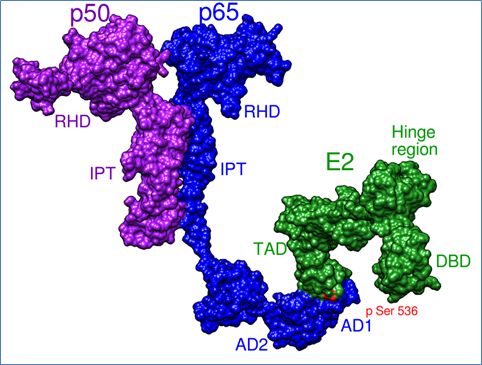
We employ molecular simulations (Modelling and Dynamics), image informatics, network and pathway mapping strategies to understand chromatin dynamics and signalling. Pharmacophore and virtual screening based drug design approaches to extract isoform specificity on epigenetic inhibitors are also extensively used.
